Brochures
NEW! Arraystar GlycoRNA Array
Discover the Pivotal Roles of Glycosylated RNAs in Cancer and Disease
• Unprecedented scientific and medical opportunities
• Cover all classes of glycoRNAs and their fragments
• Discovery of novel glycoRNAs
• Suitable for all sample types
• High sensitivity and reliability
NEW! Non-coding RNA and Epitranscriptomic Solutions
• GlycoRNA Microarrays
Cover Y-RNA/Y-RNA fragment, tRNA,tiRNA&tRF, pre-miRNA,miRNA &
more
• Downstream-of-Gene Transcript(DoG RNA) Microarrays
Profile DoG RNAs and all their target RNA types simultaneously
• R-loop Profiling
Profile the mRNA/lncRNA organized R-loops in gene regulation by
DRIPc-seq
• Small RNA Modification Microarrays
Quantify o8G/m7G/m6A/pseudouridine/m5C in miRNAs, pre-miRNAs
&tsRNAs
• Circular RNA Microarrays
Accurately profile circular RNAs by highly specific circular junction probe
design
• LncRNA Microarrays
Overcome the limitations of RNA-seq for lncRNAs often at low
abundance
• Small RNA Microarrays
Accurately profile miRNA, pre-miRNA, tRNA, tsRNA, and snoRNA
simultaneously
• Epitranscriptomic Microarrays
Quantify the percentage of m6A modification at transcript specific level
• m6A Single Nucleotide Microarrays
Locate and quantify the exact m6A site at single nucleotide resolution
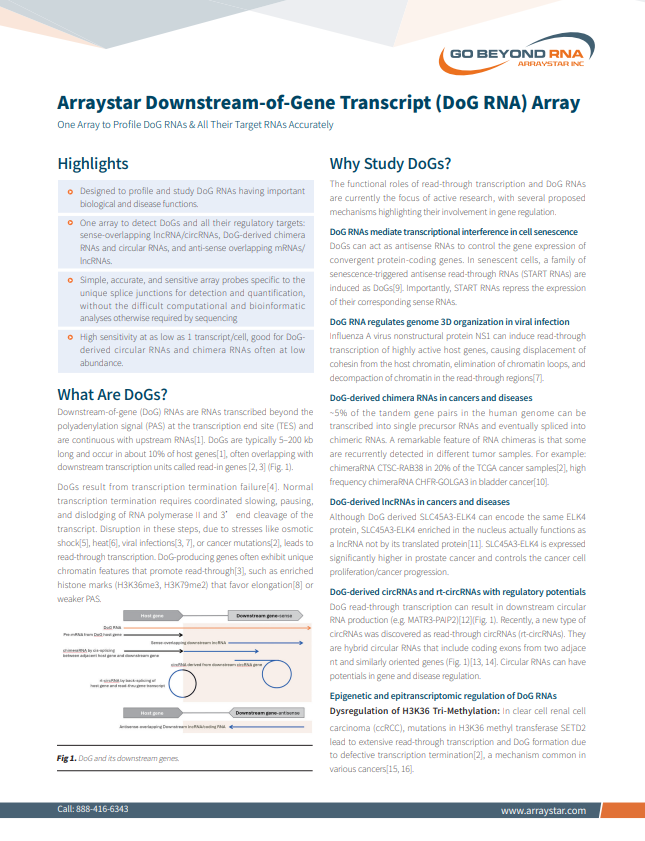
NEW! Arraystar Downstream-of-Gene Transcript (DoG RNA) Array
One Array to Profile DoG RNAs & All Their Target RNAs Accurately
• Designed to profile and study DoG RNAs having important biological and disease functions
• One array to detect DoG RNAs and all their regulatory targets
• Simple, accurate, and sensitve array probes, without the difficult computational and bioinformatic analyses otherwise required by sequencing
• High sensitivity good for DoG RNA targets often at low abundance
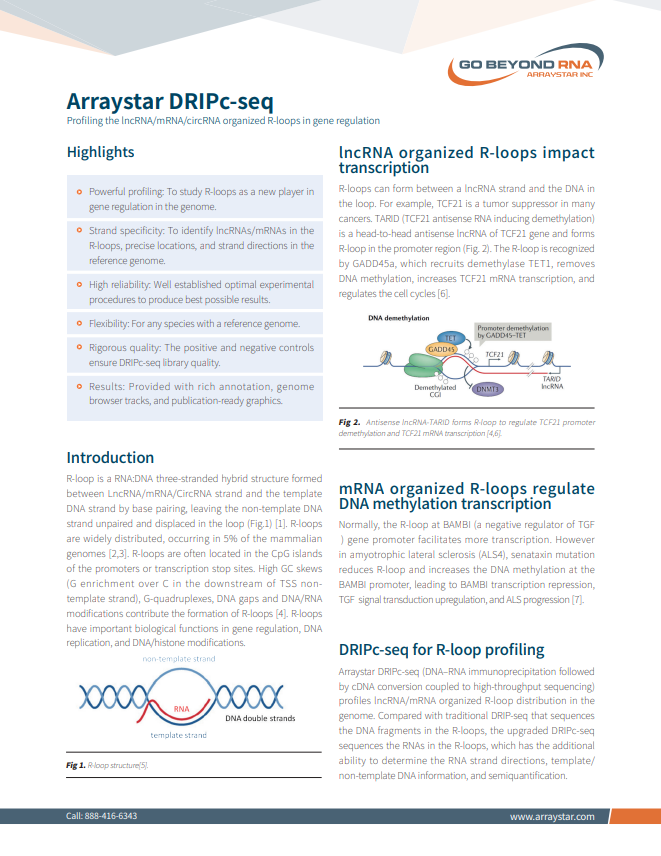
R-loop Profiling (DRIPc-Sequencing)
Profiling the LncRNA/mRNA/CircRNA Organized R-loops in Gene Regulation
• Powerful profiling: To study R-loops as a new player in gene regulation in the genome.
• Strand specificity: To identify LncRNAs/mRNAs in the R-loops, precise locations, and strand directions in the reference genome.
• High reliability: Well established optimal experimental procedures to produce best possible results.
• Rigorous quality: The positive and negative controls ensure DRIPc-seq library quality.
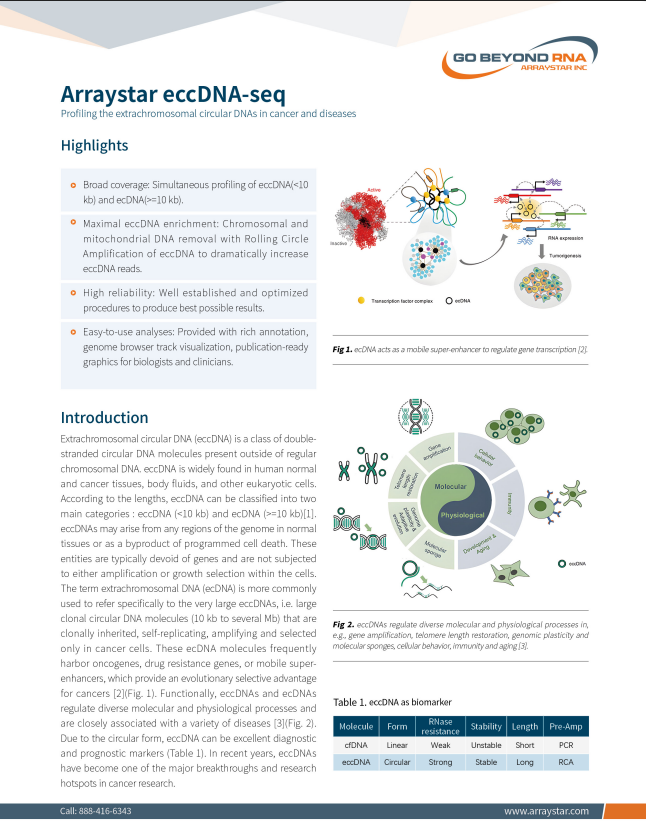
eccDNA Sequencing
Profiling the extrachromosomal circular DNAs in cancer and diseases
• Simultaneous profiling of eccDNA(<10kb) and ecDNA(>=10kb)
• Maximal eccDNA enrichment by chromosomal and mitochondrial DNA removal with Rolling Circle Amplification
• Well established and optimized procedures
• Provided with rich annotation, publication-ready graphics
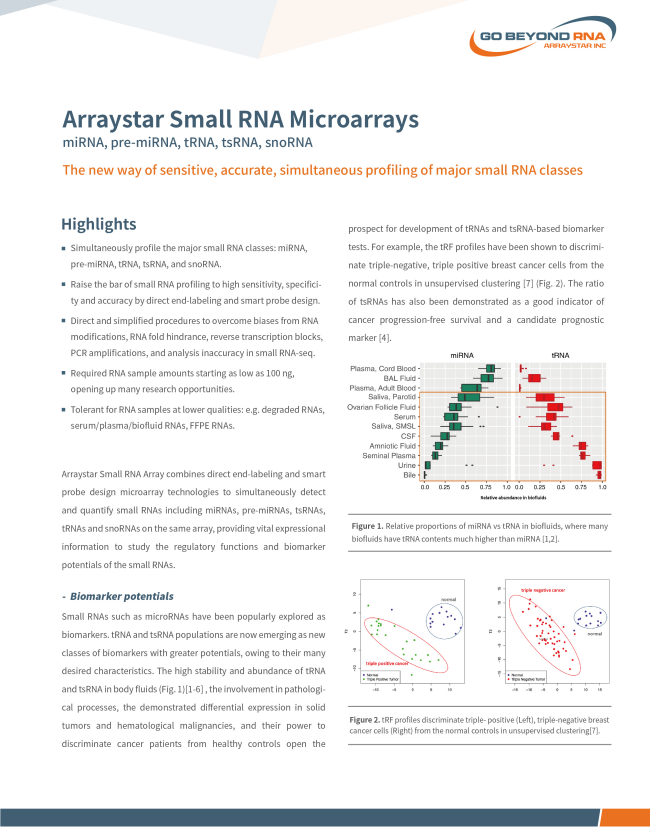
Arraystar Small RNA Array
Sensitive, accurate, simultaneous profiling of major small RNA classes
• For miRNA, pre-miRNA, tRNA, tsRNA, and snoRNA
• Sample amounts starting as low as 100ng
• Good for serum/plasma/biofluid RNAs, FFPE RNAs.
Arraystar m6A Single Nucleotide Arrays
• Single-nucleotide resolution for m6A site location
• m6A modification stoichiometry
• Low RNA sample amount requirement
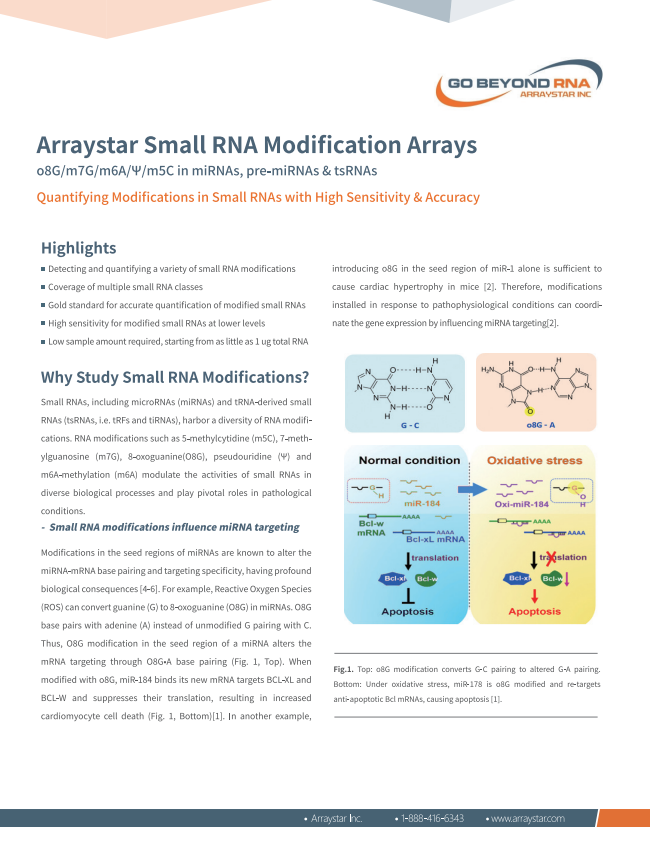
Arraystar Small RNA Modification Array
Quantifying Modifications in Small RNAs with High Sensitivity & Accuracy
– o8G/m7G/m6A/m5C in miRNAs, pre-miRNAs & tsRNAs
• Detecting and quantifying a variety of small RNA modifications
• Coverage of multiple small RNA classes
• Gold standard for accurate quantification of modified small RNAs
• High sensitivity for modified small RNAs at lower levels.
• Sample amounts starting as low as 1ug
Arraystar LncRNA Arrays
- New Versions Released in 2019
• Best for LncRNA profiling
• Full-length LncRNA collection
• Towards systematic and functional annotation of LncRNAs
• Transcript-specific detection
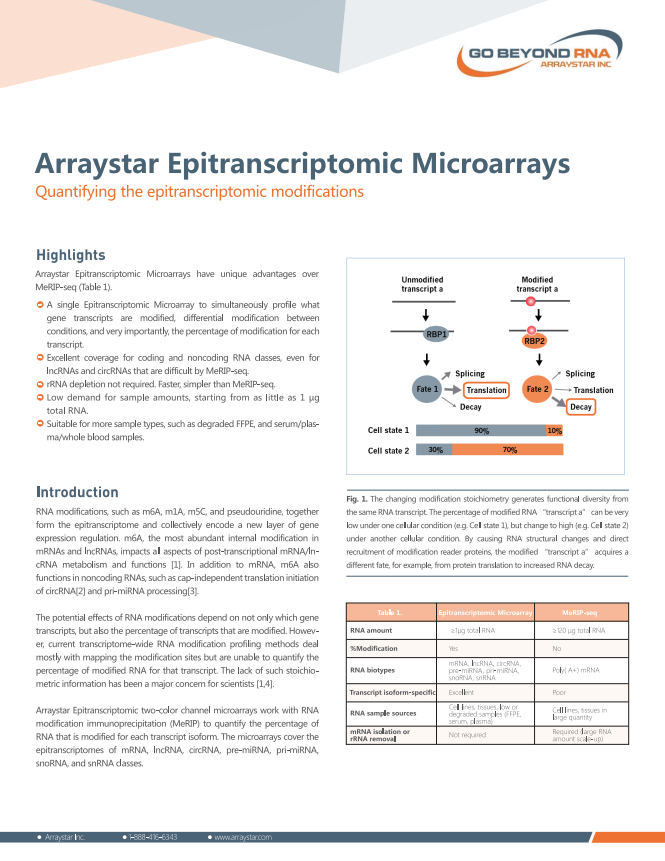
Arraystar Epitranscriptomic Microarrays
• Quantifying the percentage of modification
• Covering coding and noncoding epitranscriptomes
• Transcript isoform specific profiling
• Low sample amount requirements
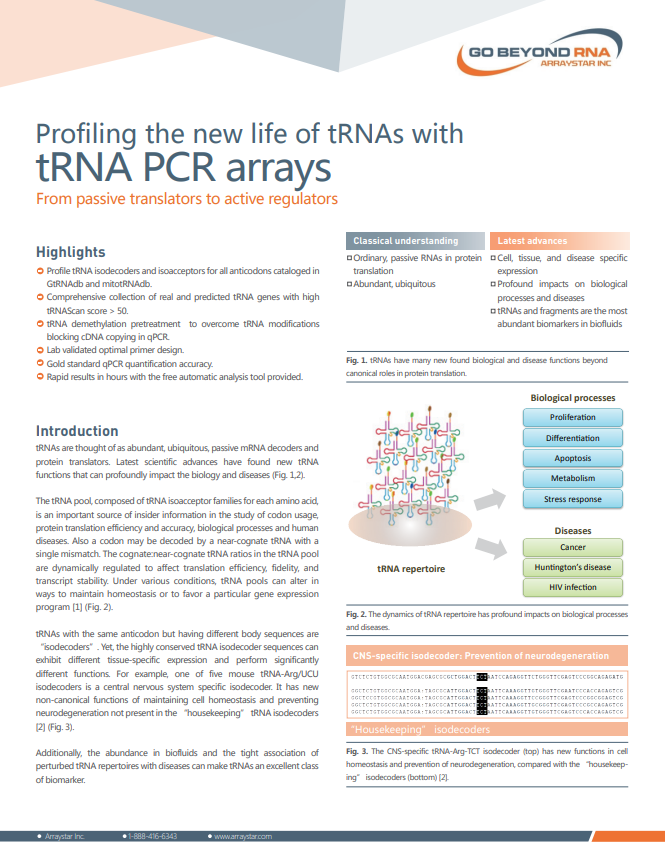
Profiling the new life of tRNAs with tRNA PCR arrays
- From passive translators to active regulators
• Isodecoders and isoacceptors
• tRNA demethylation to gain high signal sensitivity
• automatic analysis and annotation
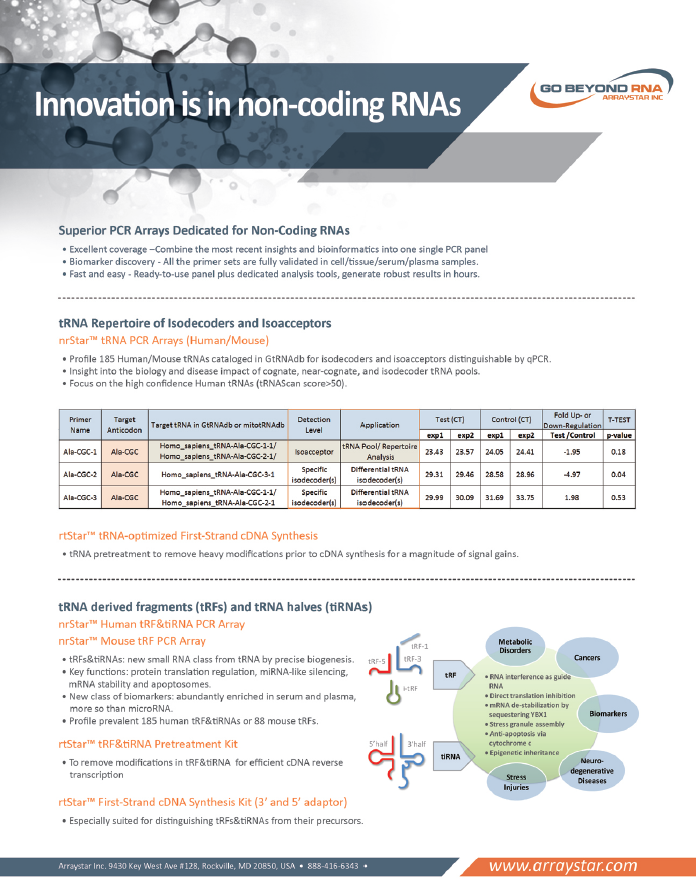
Innovation is in the non-coding RNAs
- Superior PCR Arrays Dedicated for Non-Coding RNAs
• tRNA Repertoire of isodecoders and isoacceptors
• tRNA derived fragments (tRFs) and tRNA halves (tiRNAs)
• Small Nucleolar RNAs (snoRNAs)
• Long non-coding RNAs (lncRNAs)
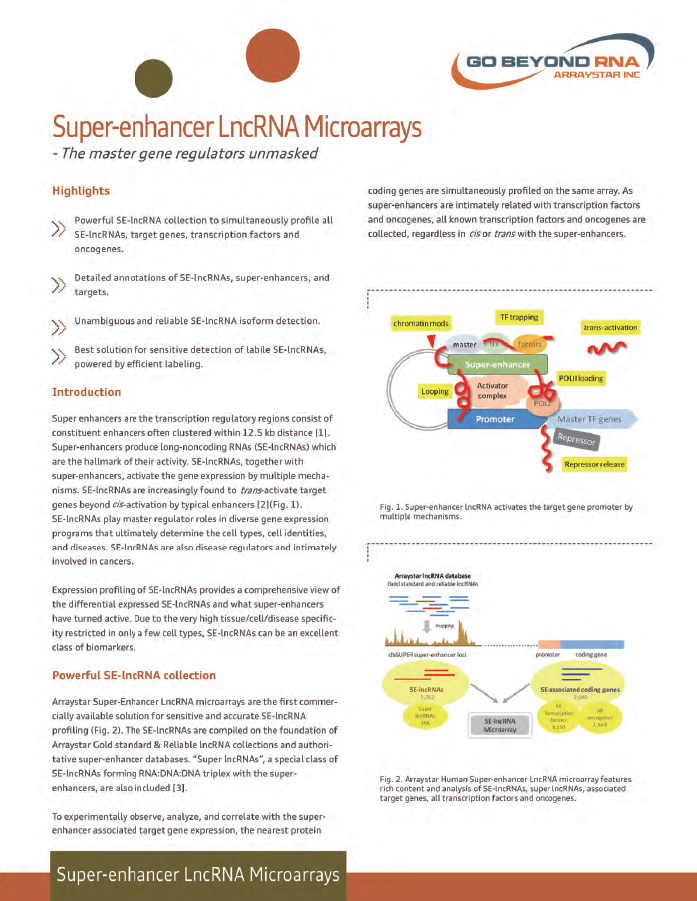
Super-enhancer LncRNA Arrays
- The master gene regulators unmasked
• Powerful SE-lncRNA collection to simultaneously profile all SE-lncRNAs, target genes, transcription factors and oncogenes.
• Detailed annotations of SE-lncRNAs, super-enhancers, and targets.
• Unambiguous and reliable SE-lncRNA isoform detection
• Best solution for sensitive detection of labile SE-lncRNAs, powered by efficient labeling.
Cutting edge research in the hottest science with NuRNA™ PCR Arrays
• NuRNA™ Human tRNA Modification Enzymes PCR Array
• NuRNA™ Human Small RNA Biogenesis Proteins PCR Array
• NuRNA™ Human Epitranscriptomics PCR Array
• NuRNA™ Human Central Metabolism PCR Array
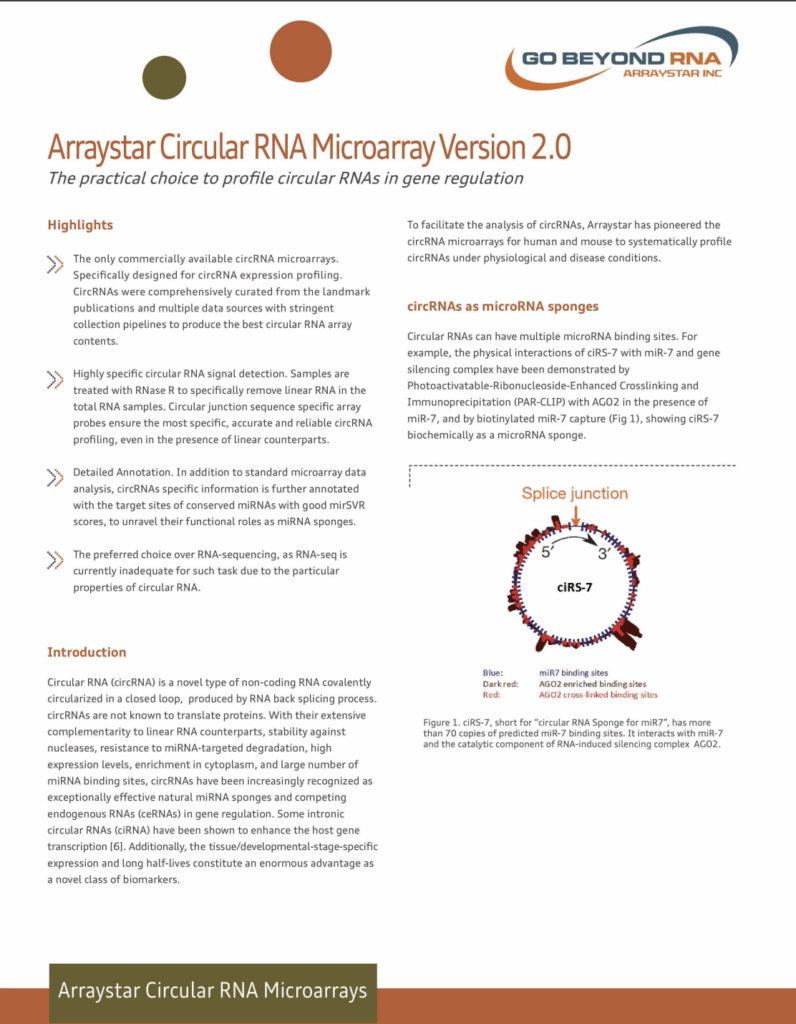
CircRNA Array V2.0
- Profile a Novel Class of RNA with Regulatory Potency
• The only commercially available circRNA microarray with the best content
• Highly specific circular RNA signal detection
• Detailed Annotation
• The preferred choice over RNA-sequencing
LncPath™ Pathway Focused LncRNA Arrays
- Discover New Players in Pathways and Diseases - LncRNAs and their target genes
• Pathway focused or disease specific
• Reliable LncRNA collection for specific pathways
• Efficient and robust labeling system
• Spike in RNA controls
• Innovative probe design
• Guaranteed performance