Epitranscriptomic Sequencing
Related Products
Related Services
Related Reviews
mim-tRNA-sequencing
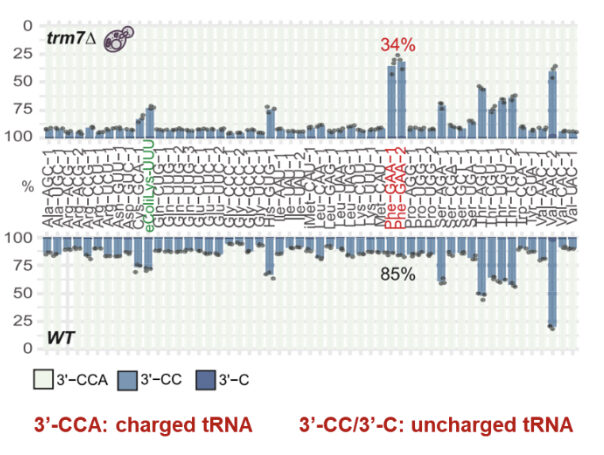
mim-tRNA-seq (modification-induced misincorporation tRNA-seq) is a novel and powerful tRNA-seq that can profile tRNA expression, tRNA modifications, and tRNA charging [12]. The method uses thermostable reverse transcriptase TGIRT to synthesize full length cDNA for the tRNAs at high efficiency, allowing quantitative analysis of the tRNA expression profiles. As tRNA modifications induce base misincorporations during TGIRT reverse transcription, mim-tRNA-seq detects the modifications at precise base positions based on the induced mutations in the tRNA sequences. By analyzing 3’-CCA end oxidation/ β-elimination, the amino acid charged or uncharged status can be measured.
mim-tRNA-seq overcomes many difficulties of reverse transcription blocks in cDNA synthesis by tRNA modifications and low full length cDNA yields with old tRNA-seq techniques. It provides tRNA abundance, tRNA modification, and tRNA charging profiles key to tRNA studies in, for example, cancer drug resistance, cardiac fibrosis, and many other diseases.
Advantages
- Simultaneous tRNA profiles: tRNA expression, tRNA modification, and tRNA charging.
- High yields for full length tRNAs: Highly efficient full length cDNA synthesis by TGIRT to reduce mapping/counting inaccuracy.
- Broad modification coverage: tRNA modifications, e.g. m1A, m1G, m3C, acp3U, are detected and quantified at single nucleotide resolution.
- Seamless integration with translatomics: To correlate tRNA charging with translation activities.
- Rich data and analyses: A wealth of tRNA multi-omics data come with common analyses (e.g. differential analyses) and detailed annotations, for comprehensive insights into the tRNAs.
- Publication-ready graphics and visualization
Integrative analyses also available with other Arraystar services : mRNA-seq, tRF&tiRNA-Seq, Arraystar Small RNA Microarray, Arraystar Small RNA Modification Microarray.
| Service Name | Description | Price |
|---|---|---|
| mim-tRNA-Seq | tRNA expression, tRNA modification, tRNA charging |
tRNAs are abundant small RNAs for carrying amino acids and decoding genetic codons for protein translation in the cells [1]. In addition to the canonical protein translation function, tRNAs have been discovered more recently as active regulators in mRNA translation and diseases by various mechanisms [2-4]. tRNA expression, tRNA modification, and tRNA charging are key profiles in regulating tRNA molecular functions (Fig.1).
tRNA expression levels profoundly impact mRNA translation. During pathogenesis, changed tRNA repertoires by differential tRNA expression directly regulate the translation efficiency and accuracy of target mRNAs by codon preferences [2]. Cell proliferation, differentiation, and apoptosis are often dynamically regulated by tRNA expression levels.
tRNA modifications exert great influence on protein translation. Certain modifications (e.g. m7G, m1A) can increase the affinity between the tRNA and ribosome to elevate mRNA translation speed and stability, whereas aberrant tRNA modifications can cause ribosome stalling or decoding errors leading to protein synthesis problems [3, 5].
tRNA charging is correlated with mRNA translation efficiency. As only the amino acid charged tRNAs can enter ribosome and pair with the anticodons, their aminoacylation levels directly determine the tRNA translation activities. That is, when the tRNA charging is low, the translation efficiency is reduced, even enough to touch off mRNA decay, cell malfunction, and pathogenesis [4, 6].
Increasingly, abnormalities in tRNA expression, modification, and charging are linked to cancer [5, 7], cardiovascular [8, 9], neurodegenerative [10], and viral infectious [11] diseases.
Despite their vital importance, profiling tRNA expression, modification, and charging had been technically difficult in the past due to highly stable tRNA fold structure and heavy tRNA modifications that hinder traditional small RNA sequencing library construction, resulted in poor cDNA yields, biased fragment coverage, and low quantitative accuracy. Arraystar min-tRNA-seq is the most advanced technology to overcome all the difficulties to simultaneously profile tRNA expression, tRNA modifications, and tRNA charging.
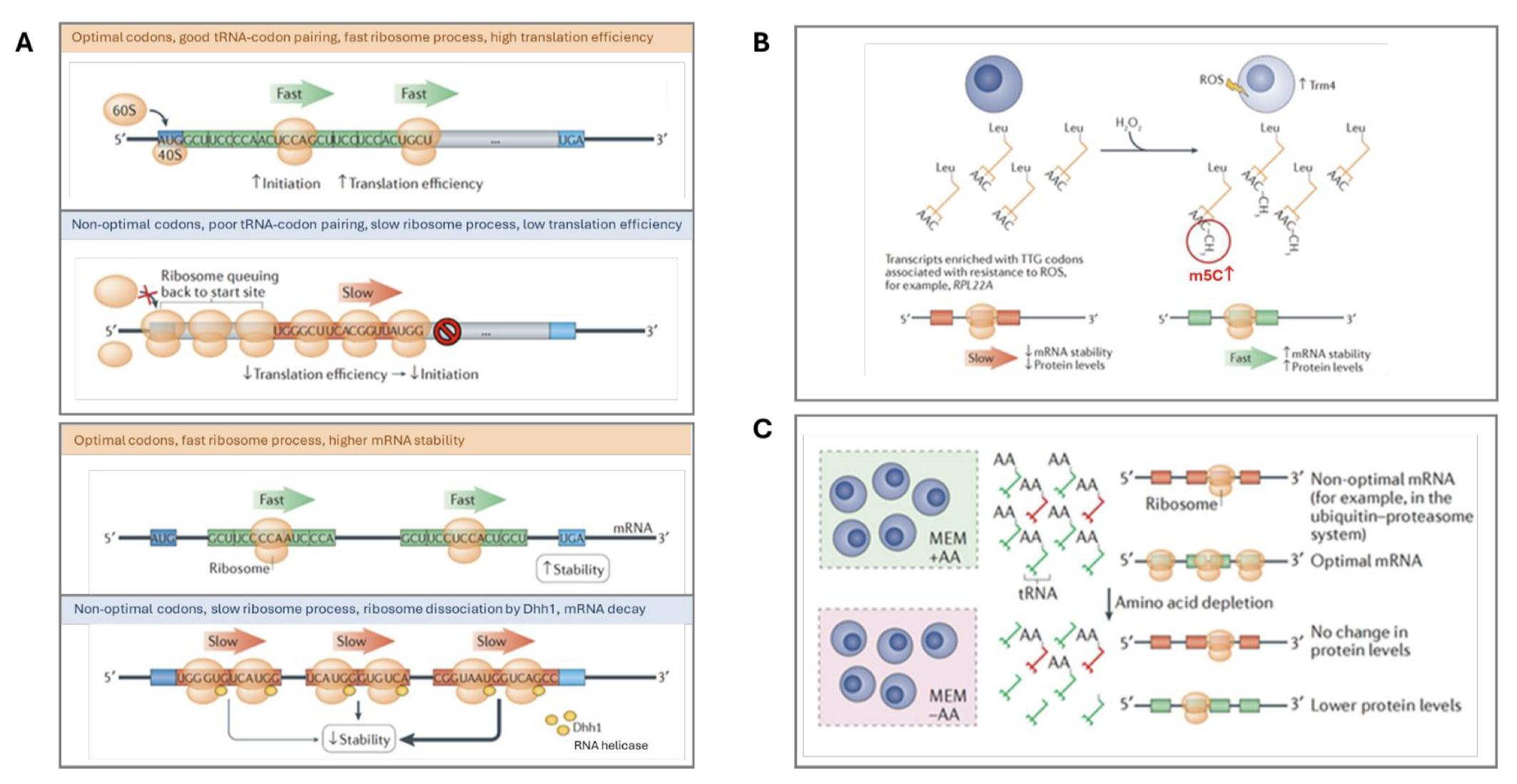
Figure 1. (A) tRNA expression on translation efficiency and mRNA stability. When an mRNA is well supplied with optimal codon tRNAs, the ribosomes progress fast and the translation is at high efficiency. When an mRNA is limitedly supplied with non-optimal codon tRNAs, the ribosomes progress slowly and the translation is at low efficiency. When mRNA is translated slowly, Dhh1 RNA helicase dissociates ribosomes from the mRNA, causing the mRNA to decay. (B) tRNA modification on translation efficiency and mRNA stability. Here, m5C modification in the tRNA anticodon increases ribosome progression speed, protein translation efficiency, and mRNA stability. The increased RPL22A protein, for example, renders the cells more resistant to reactive oxygen species (ROS). (C) tRNA charging on translation. Increased tRNA charging in cells cultured with the supplemented amino acid has higher protein translation with the optimal mRNA.
References
[1] Orellana, E.A., E. Siegal, and R.I. Gregory, tRNA dysregulation and disease. Nature Reviews Genetics, 2022. 23(11): p. 651-664.
[2] El-Hachem, N., et al., Valine aminoacyl-tRNA synthetase promotes therapy resistance in melanoma. Nat Cell Biol, 2024. 26(7): p. 1154-1164.
[3] Dai, Z., et al., N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell, 2021. 81(16): p. 3339-3355.e8.
[4] Wu, J., et al., Glutamyl-Prolyl-tRNA Synthetase Regulates Proline-Rich Pro-Fibrotic Protein Synthesis During Cardiac Fibrosis. Circ Res, 2020. 127(6): p. 827-846.
[5] Qi, L., et al., Dual Targeting of m(7)G tRNA Modification and Histone Acetylation using Carrier-Free Nano-Epidrugs to Evoke Osteosarcoma Chemosensitization. Adv Mater, 2025: p. e05951.
[6] Duan, Y., et al., A glutamyl-tRNA reductase and its binding protein promote transitory starch biosynthesis and enhance grain quality and yield in rice. Plant Commun, 2025. 6(11): p. 101527.
[7] Qian, Y., et al., ALKBH8-mediated codon-specific translation promotes colorectal tumorigenesis. Nat Commun, 2025. 16(1): p. 9075.
[8] Nah, J., et al., Microprotein SMIM26 drives oxidative metabolism via serine-responsive mitochondrial translation. Mol Cell, 2025. 85(14): p. 2759-2775.e12.
[9] Das, A.S., et al., AIMP3 maintains cardiac homeostasis by regulating the editing activity of methionyl-tRNA synthetase. Nat Cardiovasc Res, 2025. 4(7): p. 876-890.
[10] Beharry, A., et al., High-fidelity and differential nonsense suppression in live cells and a frontotemporal dementia allele with human transfer RNAs. Nucleic Acids Res, 2025. 53(14).
[11] Yang, X., et al., Anticodon Engineered Transfer RNA (tRNA(SUAG)) Inhibits Hepatitis B Virus Replication by Promoting the Degradation of Core Protein. Adv Sci (Weinh), 2025: p. e03534.
[12] Behrens, A., G. Rodschinka, and D.D. Nedialkova, High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell, 2021. 81(8): p. 1802-1815.e7.
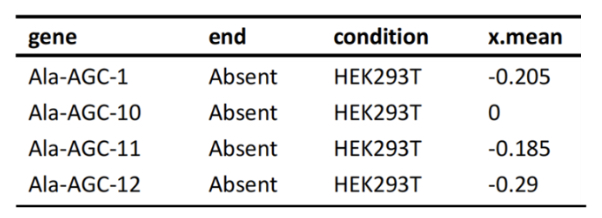
tRNA Differential Expression Analysis
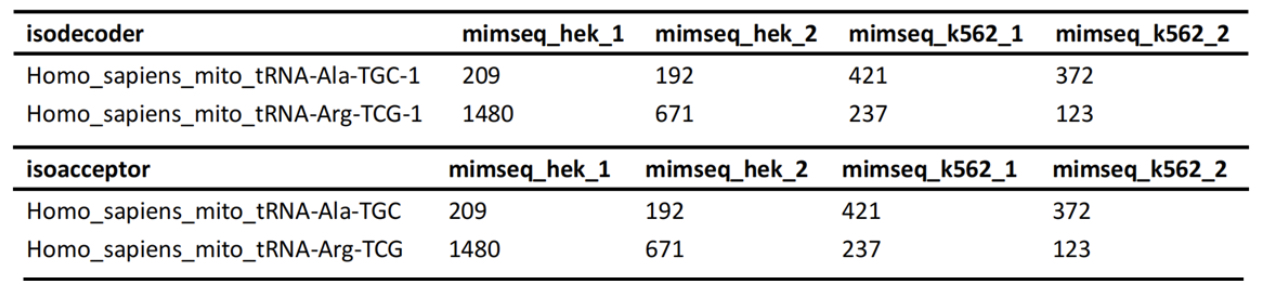
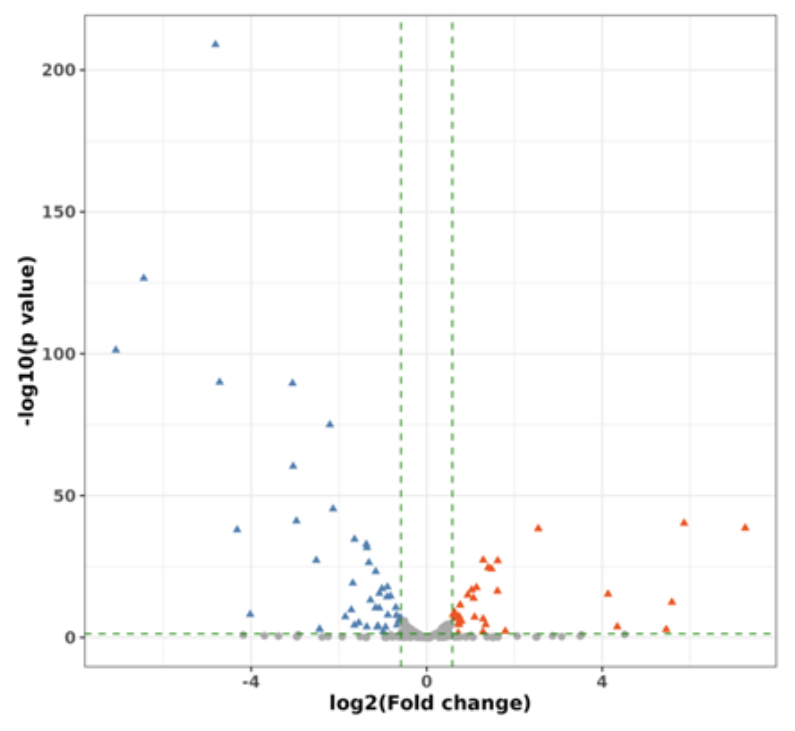
tRNA Differential Modification Analysis
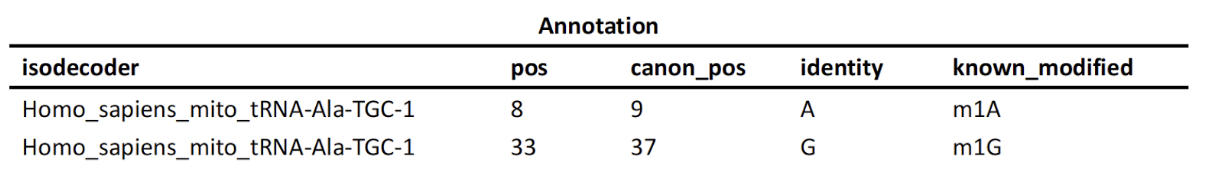
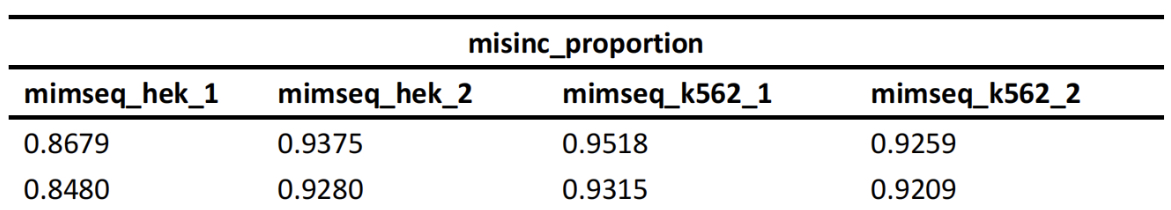
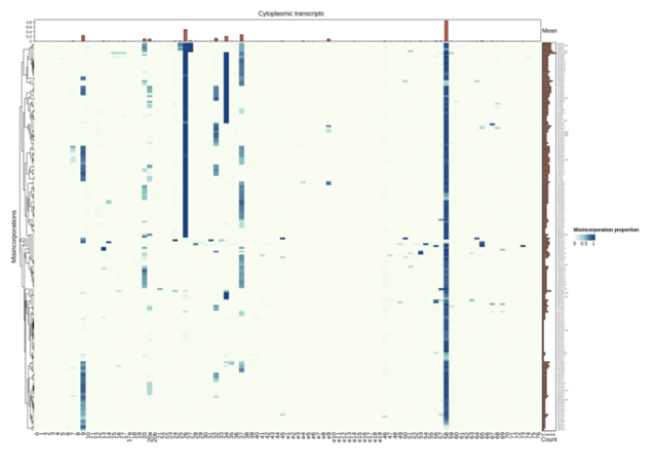
tRNA Modification Heatmap
Differential tRNA Charging Analysis