|
Customer Success Story
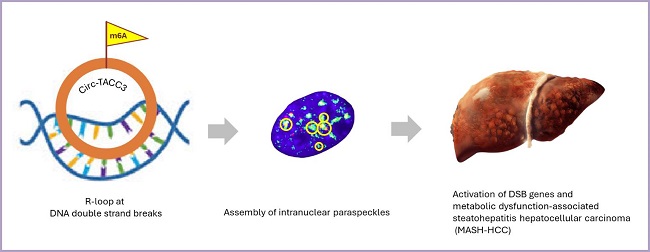
The landscape of m6A epitranscriptomic modification of circRNAs in liver cancer cells was profiled by Arraystar CircRNA Epitranscriptomic Array [1](Fig.1). circTACC3 was found the most prevalent m6A modified circRNA in metabolic dysfunction-associated steatohepatitis related hepatocellular carcinoma (MASH-HCC), which impacts on the intracellular lipid accumulation, growth, and environmental adaptive survival of tumor cells. circTACC3 assembles intranuclear paraspeckle in m6A-dependent manner, leading to chromatin remodeling and gene activation in malignant progression.
References
[1] Fu J. et al (2025) "Intranuclear paraspeckle-circular RNA TACC3 assembly forms RNA-DNA hybrids to facilitate MASH-related hepatocellular carcinoma growth in an m(6)A-dependent manner." Cancer Commun (Lond) 45(11):1583-1610 [PMID:41103024]
|
|
Arraystar Epitranscriptomic Arrays
Quantify choice of m6A/m5C/m1A/ac4C/m7G/Ψ epitranscriptomic modification in mRNAs & lncRNAs or circRNAs, with unique advantages over MeRIP-seq:
• A single Epitranscriptomic Microarray to simultaneously profile what gene transcripts are modified, differential modification between conditions, and very importantly, the percentage of modification for each transcript.
• Excellent coverage even for lncRNAs and circRNAs that are difficult to profile by MeRIP-seq.
• rRNA depletion not required. Faster, simpler than MeRIP-seq.
• Low sample amount required, starting from as little as 1 ug total RNA for mRNAs & LncRNAs or 3 ug for CircRNAs.
|