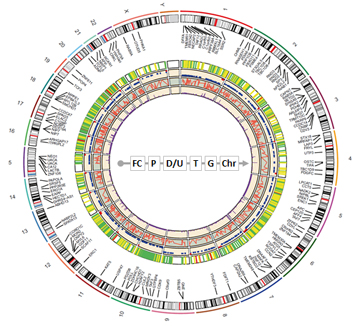
Fig 1.Circos diagram of differentially m6A-methylated sites in the jaw bone marrow mesenchymal stem cells (BMSC) of Type 2 diabetes. The circular tracks for fold change (FC), p-value (P), down/up regulation (D/U), sites in 5', 3’, CDS transcript unit (T), gene (G), and chromosome (Chr) are indicated.
|
|
Customer Success Story
Using Arraystar m6A Single Nucleotide Microarray, the m6A RNA modification was profiled at single base resolution in the jaw bone marrow mesenchymal stem cells (BMSC) with and without Type 2 diabetes [1]. Most of the differentially modified RNAs are enriched in metabolic processes. Confirmed by MazF-qPCR, the differentially modified EPB41L3, ADNP, GDF11, and RGS2 RNA transcripts have the potentials to enhance BMSC function and improve dental implantation success in T2DM patients.
Arraystar m6A Single Nucleotide Arrays
• Orthogonal m6A profiling by MazF enzyme, independent of m6A antibody based MeRIP.
• Single-nucleotide resolution for m6A sites.
• Stoichiometry for the fraction or percentage of m6A modification.
• RNA sample amount as low as 1 ug total RNA.
• Reliable m6A site collection and systematic annotations.
|

