Overcome the Challenges of Small RNA Profiling
|
| Small RNAs include miRNAs, pre-miRNAs, tsRNAs, tRNAs and snoRNAs. Even though RNA-seq has been used for profiling small RNAs, we need to overcome several major challenges unique to these small RNA classes in order to accurately profile their whole spectrum, especially for samples at limited amount. |
• Interference from RNA Modifications
Small RNA modifications (e.g. m1A, m3C and m1G, particularly heavily in tRNAs and tsRNAs) block reverse transcription, causing failures in constructing full length unskewed representation of the sequencing library.
• Library Amplification Biases
RNA G/C content, sequence contexts, secondary structures, RNA lengths, priming, and reaction conditions can lead to biases and distortions in the PCR amplification of the sequencing library [1].
• Distortions in Small RNA-seq Data Analysis using TPM
TPM, a commonly used measure in small RNA-seq data analysis, is dependent on the composition of the RNA population in a sample [2,3](Fig 1).
• Sample Amounts Required
For small RNA-sequencing of tRNA and tsRNA, 5ug or even 100ug total RNA is required for target RNA isolation and pretreatments [5], which precludes research projects with limited sample amount availability.
|
|
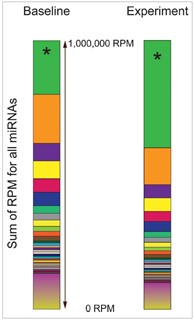
Fig 1. A few very highly expressed
genes can skew the distribution of
TPM expression values [4].
|
|
| Arraystar Small RNA Array is designed to overcome these challenges to profile the full spectrum of small RNAs at high sensitivity and accuracy, yet requiring much less sample RNA amounts. It combines direct end-labeling and smart probe design microarray technologies to detect and quantify miRNAs, pre-miRNAs, tsRNAs, tRNAs and snoRNAs simultaneously on the same array, opening up more opportunities for exploring their biomarker potentials. |
Subscribe to Our Newsletters Now!
References
[1] Dabney J. and Meyer M. (2012) Biotechniques [PMID:22313406]
[2] Zhao, Y., et al. (2021) J Transl Med [PMID: 34158060]
[3] Zhao, S., et al. (2020) RNA [PMID: 32284352]
[4] Witwer, K. W. and Halushka, M. K. (2016) RNA Biol [PMID: 27645402]
[5] Junchao S., et al. (2021) Nat Cell Biol [PMID: 33927371]
|
| Latest Publications
Circular RNA
• Yuan Q, et al. (2023)
Signal Transduction &Targeted Therapy
[PMID: 36882410]
• Zhong GL, et al. (2023)
Molecular Cancer
[PMID: 37004067]
RNA Modification
• Yan W, et al. (2023)
International Journal of Oral Science
[PMID: 36631441]
|
|
New Service
DRIPc-Seq provides valuable functional insights in transcriptional regulation by LncRNA/mRNA organized R-loops.
eccDNA Seq
not only dramatically increases eccDNA reads, but also simultaneously profiles both of micro-eccDNA and mega-eccDNA, accelerating your biomarker discovery and cancer research.
|
|
New Webinar
The Latest Highlights on
CircRNA in Cancer
Watch Video
Promotion
15% OFF CircRNA Array
Accurate CircRNA Profiling with Rich
Analysis
Valid 06/01/2023 - 08/31/2023
Request Quote
|
|
|

