|
• Interference from RNA Modifications
Small RNA modifications (e.g. m1A, m3C and m1G, particularly heavily in tRNAs and tsRNAs) block reverse transcription, causing failures in constructing full length unskewed representation of the sequencing library.
• Library Amplification Biases
RNA G/C content, sequence contexts, secondary structures, RNA lengths, priming, and reaction conditions can lead to biases and distortions in the PCR amplification of the sequencing library [1].
• Distortions in Small RNA-seq Data Analysis using TPM
TPM, a commonly used measure in small RNA-seq data analysis, is dependent on the composition of the RNA population in a sample [2,3](Fig 1).
• Sample Amounts Required
For small RNA-sequencing of tRNA and tsRNA, 5ug or even 100ug total RNA is required for target RNA isolation and pretreatments [5], which precludes research projects with limited sample amount availability.
|
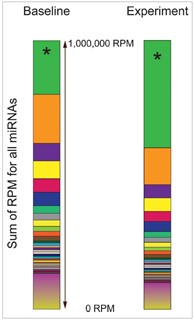
Fig 1. A few very highly expressed
genes can skew the distribution of
TPM expression values [4].
|
