Profiling m6A at single nucleotide resolution has been challenging. Conventional methods such as m6A/MeRIP-seq cannot precisely identify which adenosines in a MeRIP-seq peak are actually modified, nor can they quantify the modification fraction for each site [1].
To address these challenges, Arraystar has developed m6A Single Nucleotide Arrays based on methyl-sensitive MazF RNase that precisely locate the m6A modification at exact adenosine and quantify the stoichiometry of m6A modification fractions (Fig.1).
The arrays add invaluable benefits which are lacking for conventional methods:
- Systematic m6A profiling independent of m6A-antibody immunoprecipitation based approaches.
- Precise detection of m6ACA at single nucleotide resolution.
- Quantifying m6A modification stoichiometry and abundance.
- Low demand for sample amounts, as low as 1 ug total RNA.
- Specialized pipeline to collect and annotate the quantifiable Single-, Poly-, or Clustered-m6ACA sites.
|
|
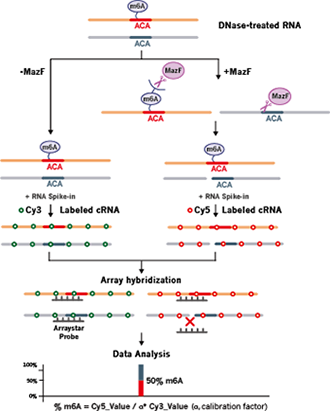
Fig 1. The workflow of Arraystar m6A Single Nucleotide Array |
Reference:
[1] Garcia-Campos M A, Edelheit S, Toth U, et al. Deciphering the "m(6)A Code" via Antibody-Independent Quantitative Profiling. Cell, 2019,178(3):731-747. |
